Technologies
Experimental Approaches:
1) Genetic modification of neural progenitors.
The lab has expertise in a number of techniques
to manipulate retinal progenitors both
in vivo and in
culture, including mouse genetics, retroviral transduction, and
electroporation. Developing retinas can be explanted and grown in tissue
culture, which facilitates a variety of experimental approaches.
Cone derived from progenitors
transfected with green fluorescent protein at embryonic day 14.5 via
in utero retinal
electroporation, and allowed to develop until postnatal day 21. Cone opsin
(cyan) and peanut agglutinin (magenta) mark the cone photoreceptors.
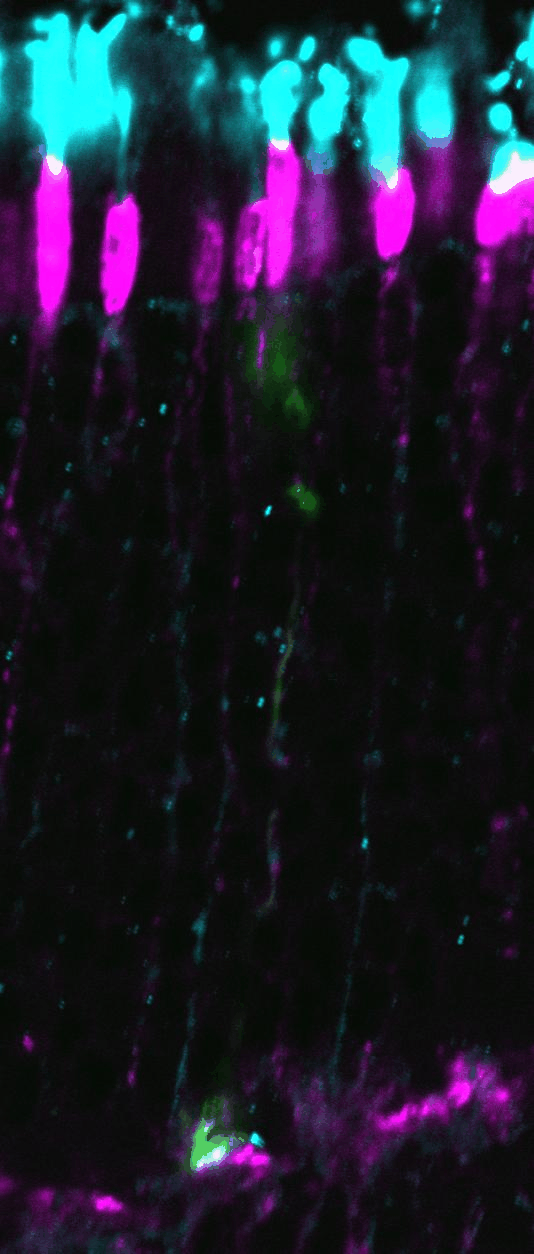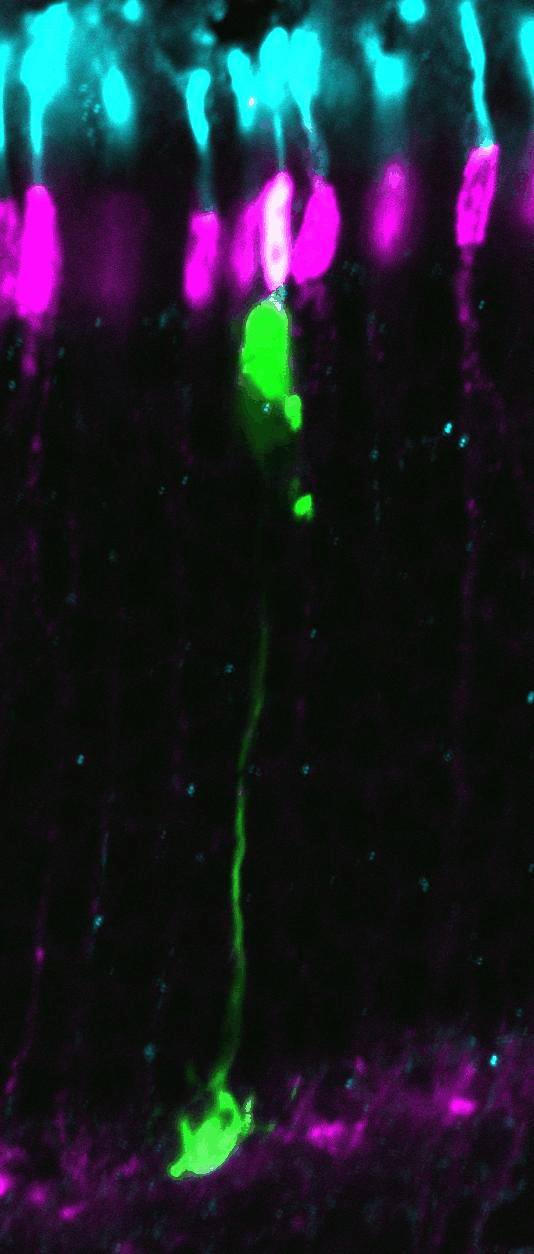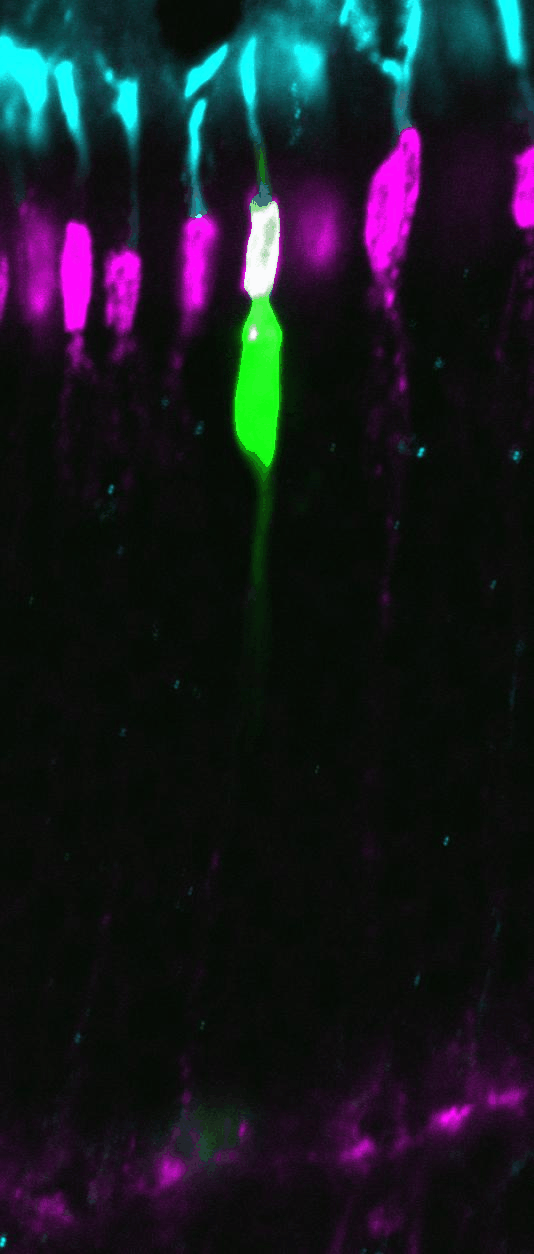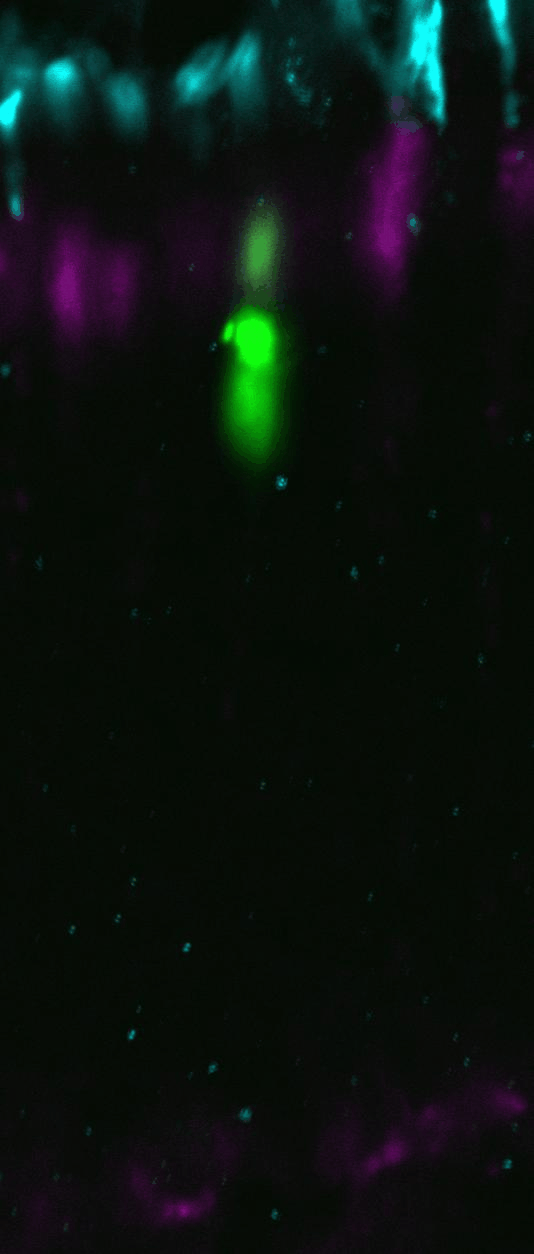
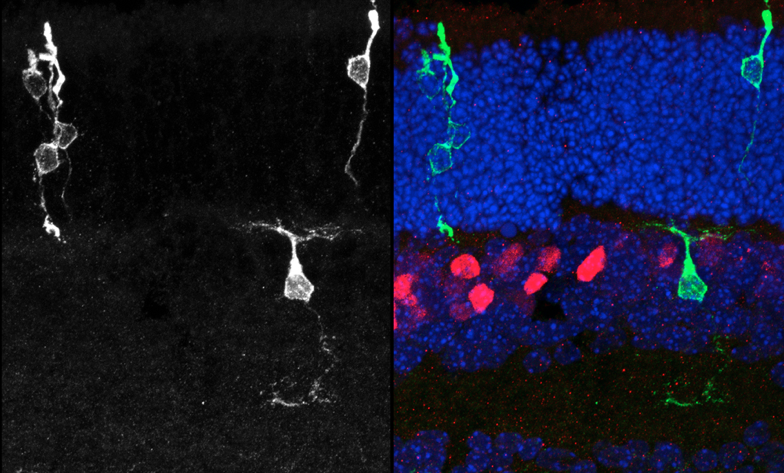
Clonal analysis allows the
complete lineage generated by individual progenitor cells to be analyzed. Here,
retroviral clones (green) generated from dividing progenitors transduced at
postnatal day 0, and harvested 2 weeks later. Vsx2 protein staining marks
bipolar cells (red), while Hoechst stains the DNA (blue), allowing the tissue
to be visualized in full. The clone on the left contains 3 rods, while the
clone on the right contains a rod and a bipolar cell.
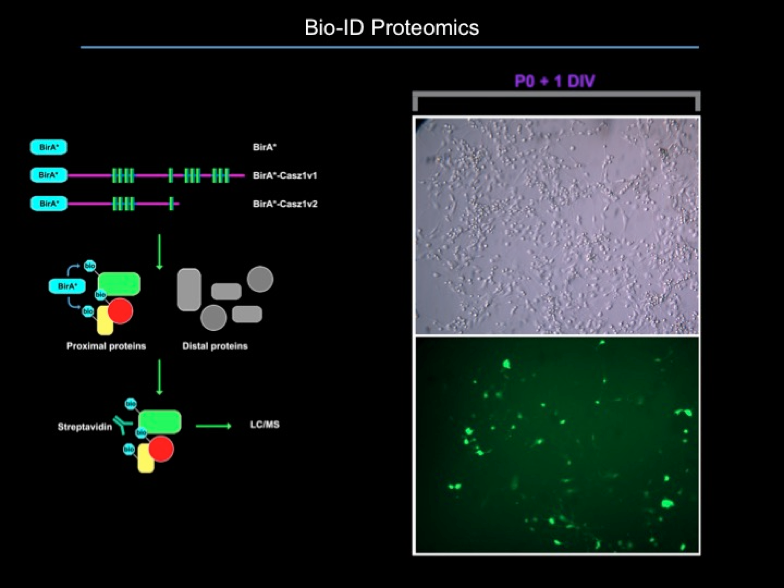
 2. Proteomics
2. Proteomics
We perform Bio-ID on primary retinal cultures, as well as co-IP/MS on tissue.
 3. Genomics and transcriptomics
3. Genomics and transcriptomics
We use cut&run-seq and ATAC-seq to examine how nucleosome remodelling complexes interact with the genome.
We use multi-seq
Multi-seq to perform multiplexed single cell RNA-seq. See
Clemot-Dupont et al.