Habibi Lab
Team Leader

Ehsan Habibi
Scientist, Regenerative MedicineWhat We Do
Research Activities
Our research builds on a unifying, multiscale–multimodal approach to decode, reconstruct, and reprogram self-organizing multicellular systems through four iterative pillars:
1. Detailed Multimodal Molecular Dissection of Embryonic Development
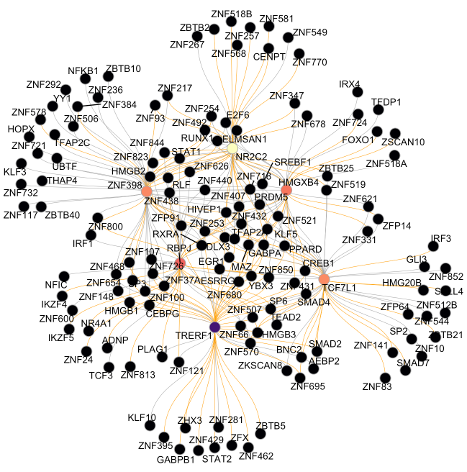
We deploy cutting-edge single-cell and spatial genomics—scRNA-seq, scATAC-seq, in situ sequencing, spatial transcriptomics—and complementary epigenomic assays to deconvolve the transcriptional and epigenetic landscapes that underlie cell-fate emergence in mouse and human embryos. By reconstructing gene-regulatory networks and trajectories and mapping the precise sequence and timing of key transcriptional and epigenetic events, we identify the molecular programs that drive symmetry breaking and lineage specification.
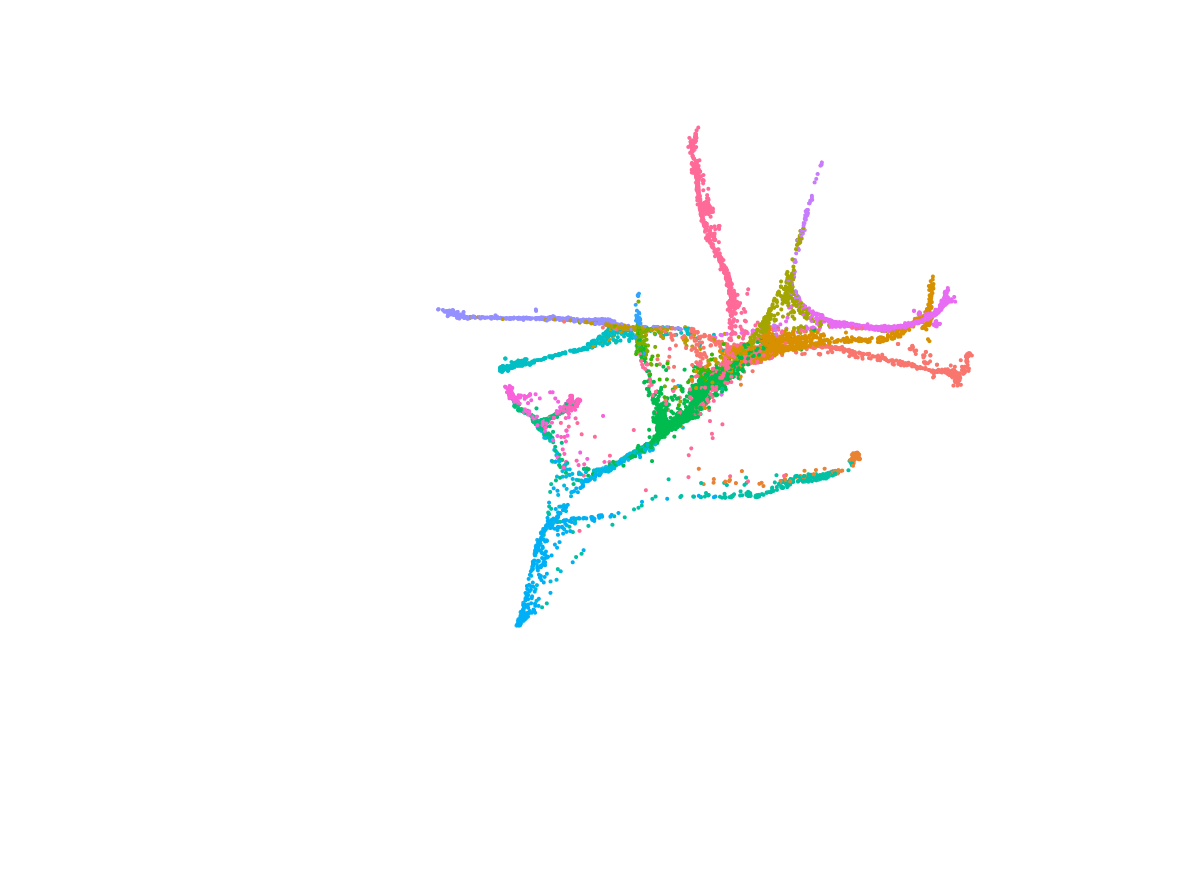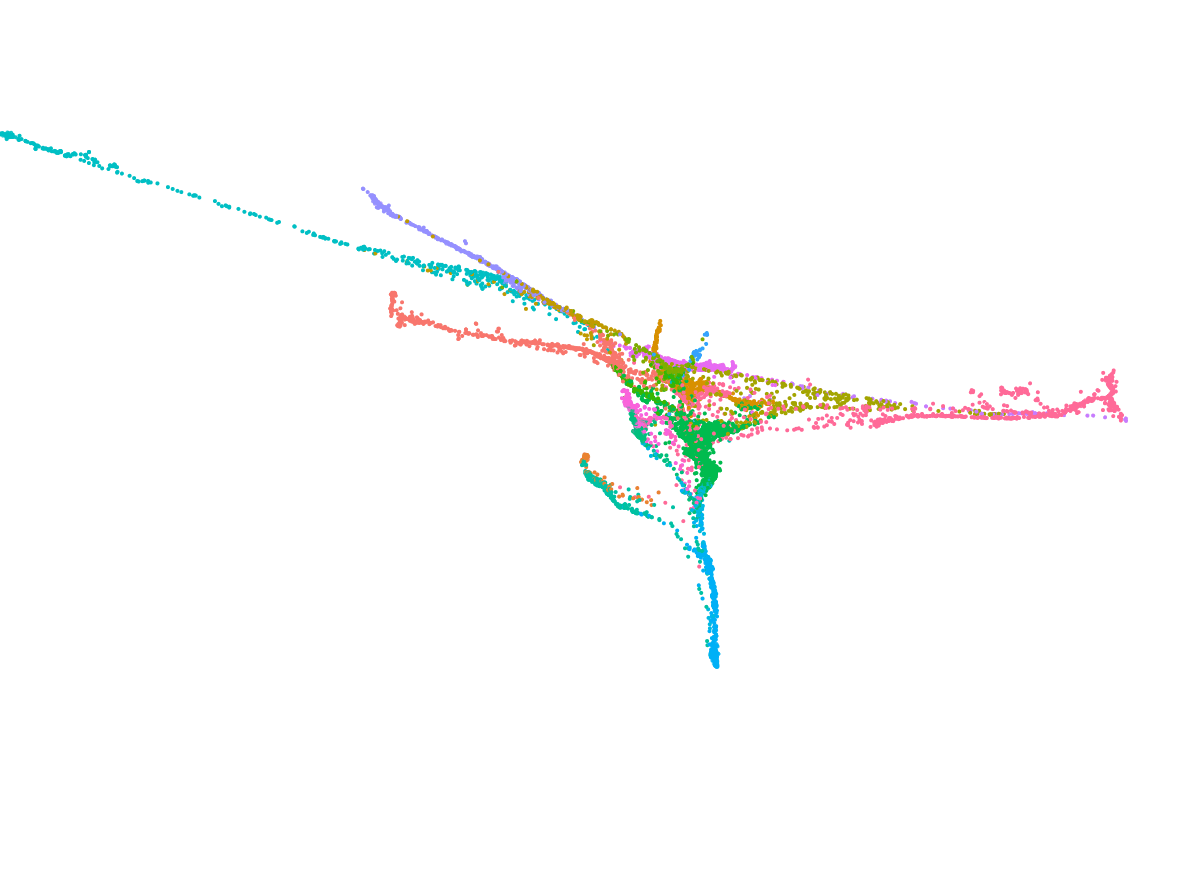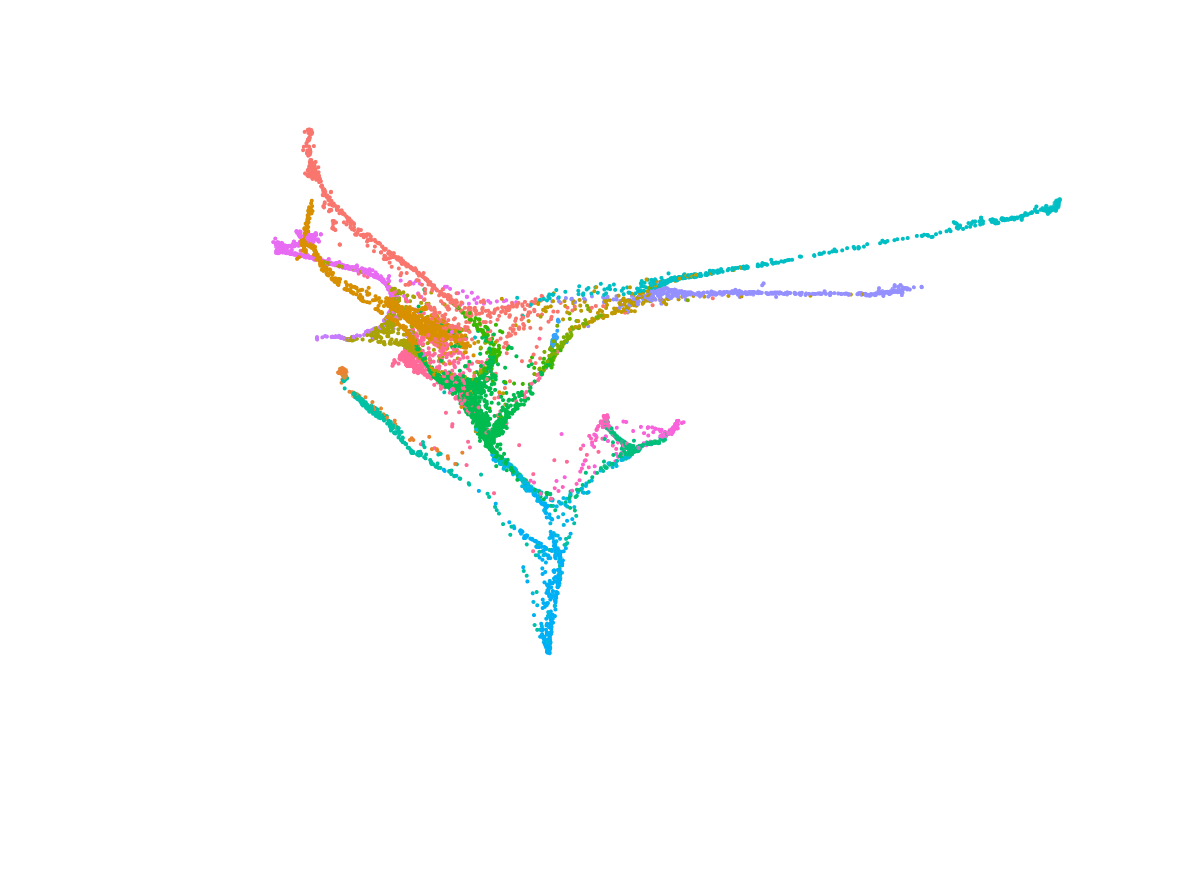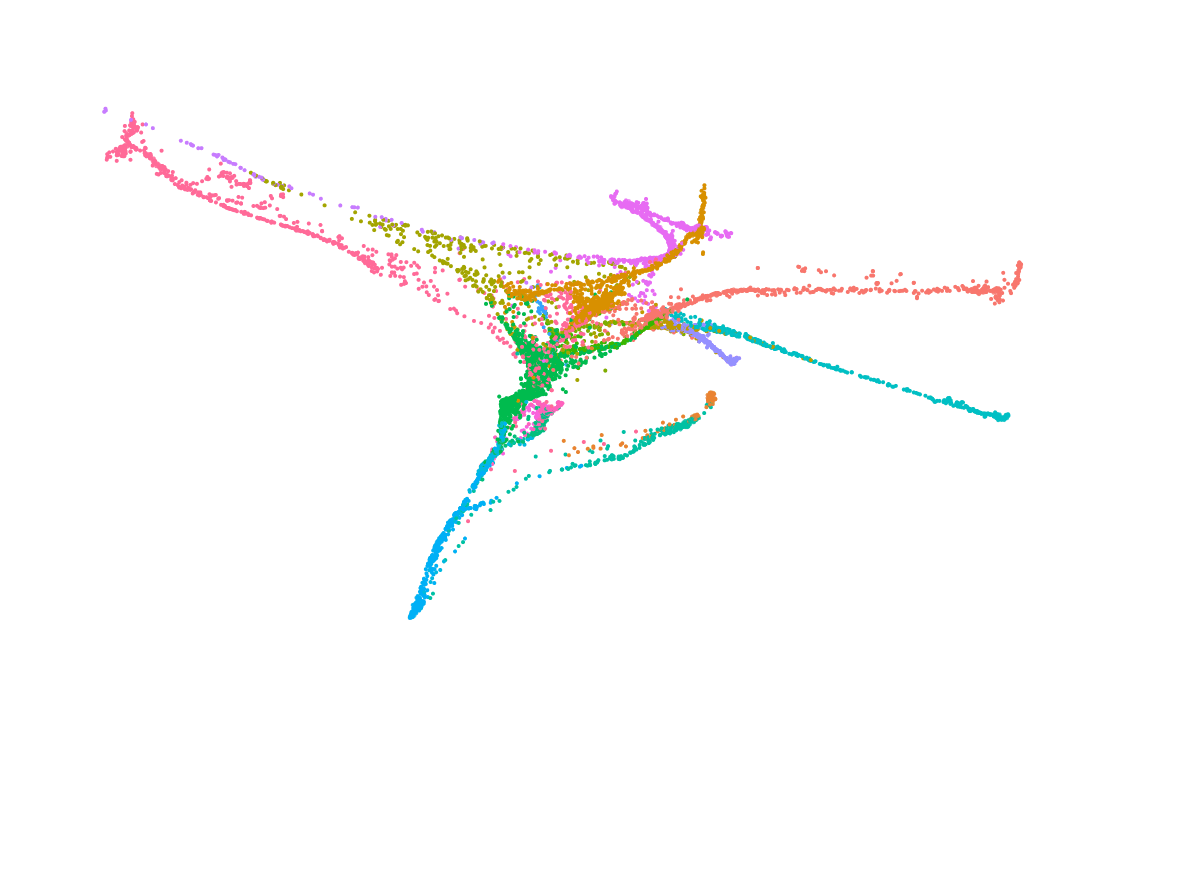
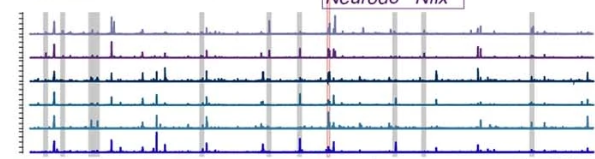
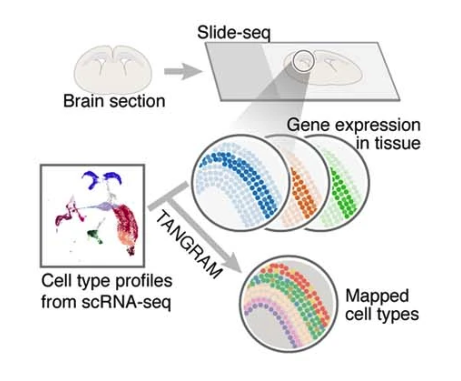
Transcriptomics
2. Developing Novel Multiscale In Situ Measurement Technologies
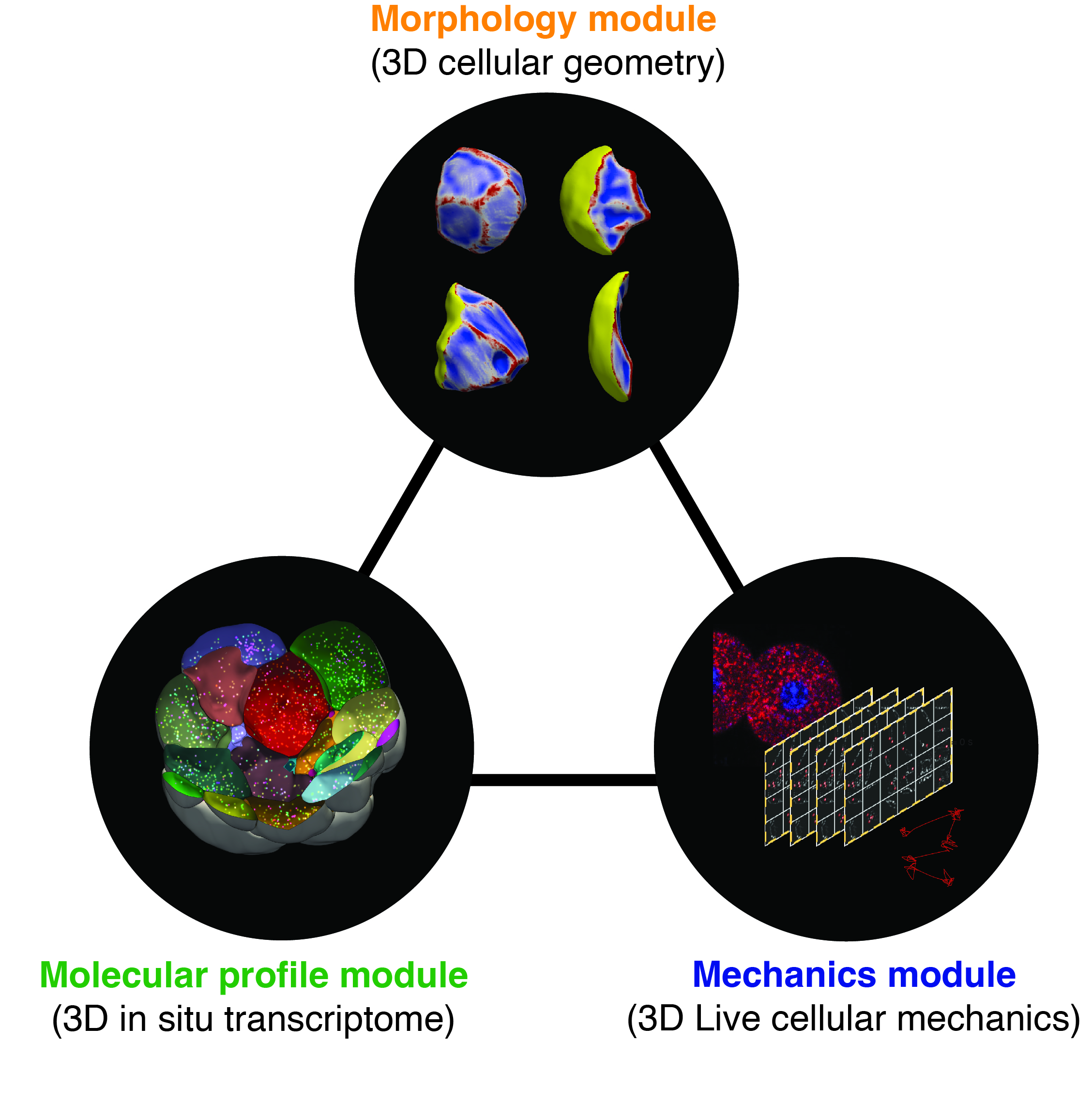
Self-organization transcends gene expression alone. To capture the full suite of morphogenetic inputs, we have developed a Unified Transcriptome and Mechanics and Morphology (UTMM) framework, which simultaneously profiles 3D spatial transcriptomes, cellular mechanics, and 3D cell geometry at single-cell resolution in intact embryos. We are extending UTMM to include other modalities—providing a truly integrative view of how molecular, physical, and geometric cues co-evolve across space and time.
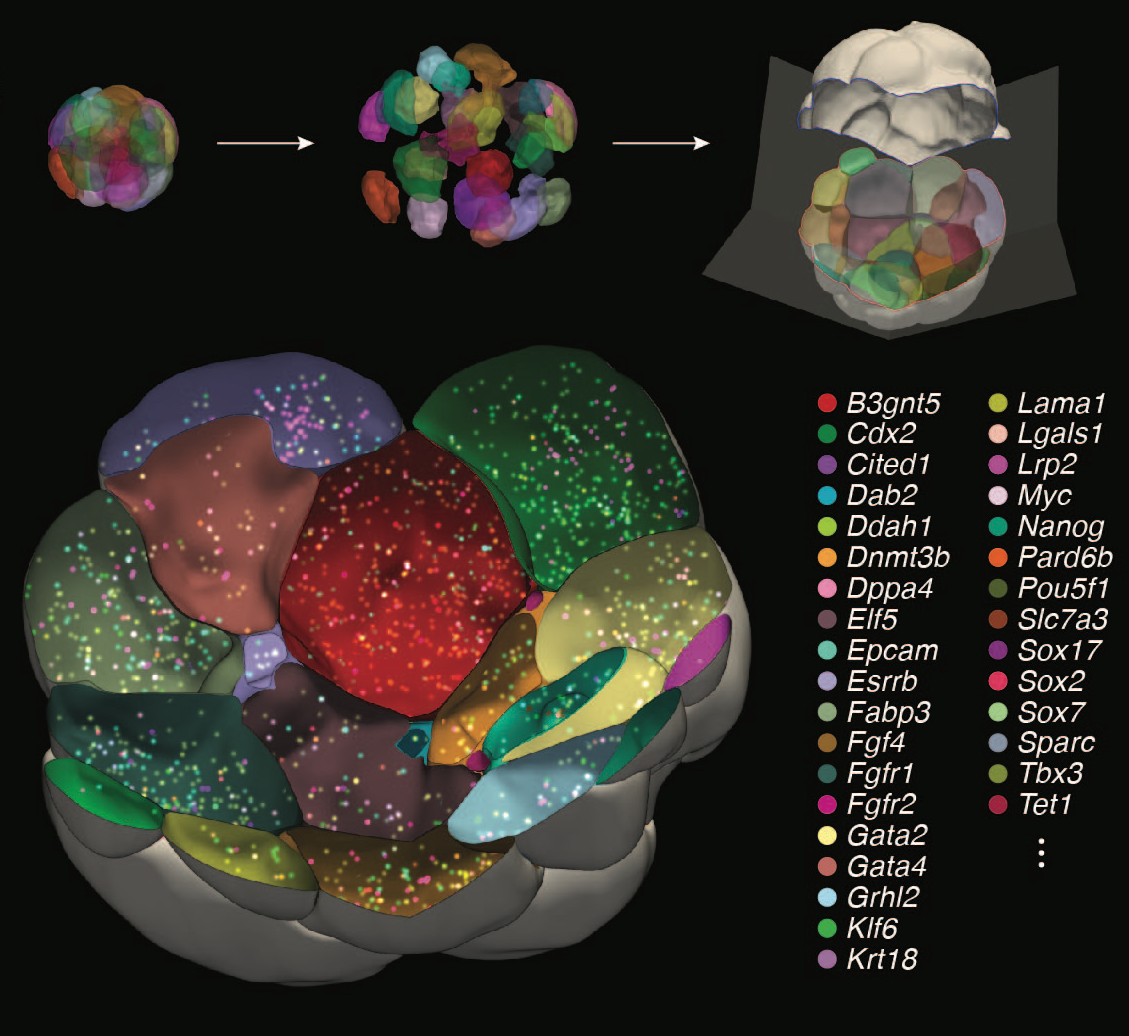
Transcriptome
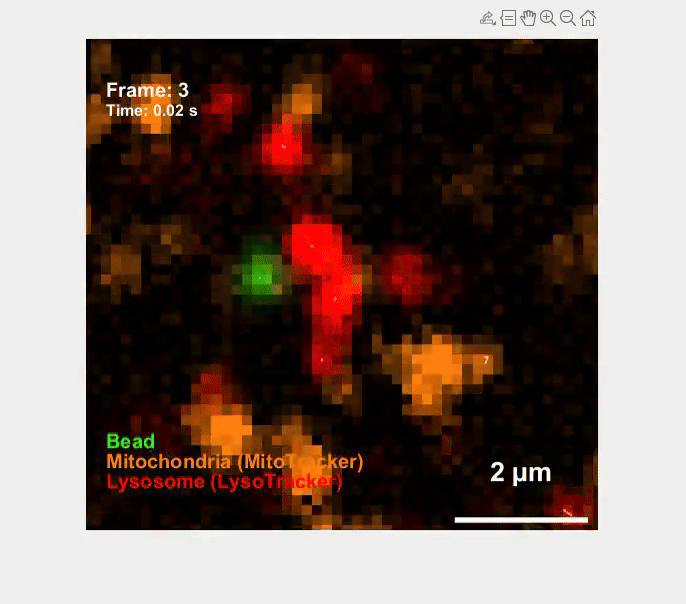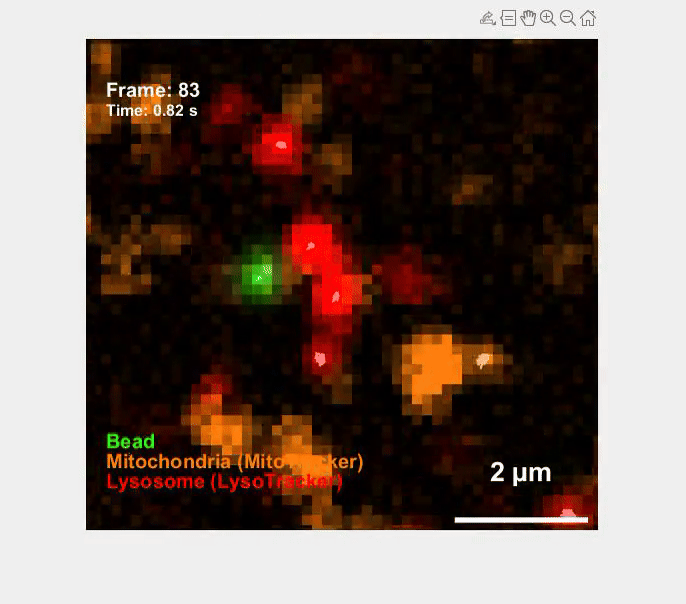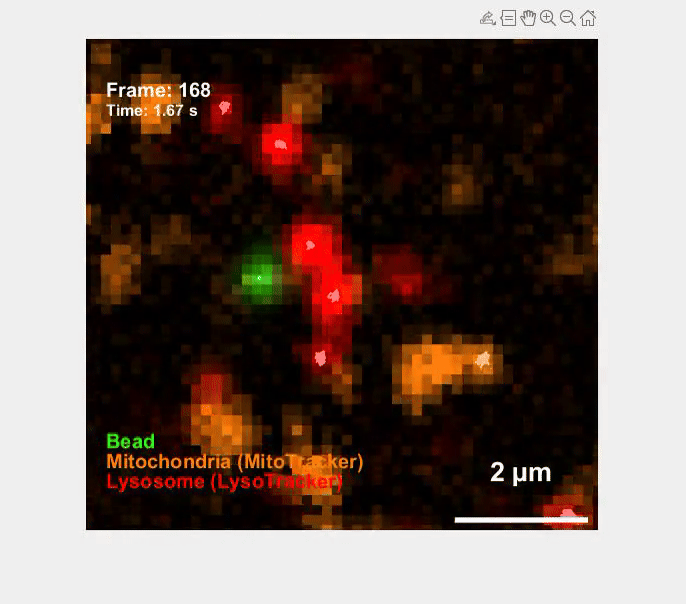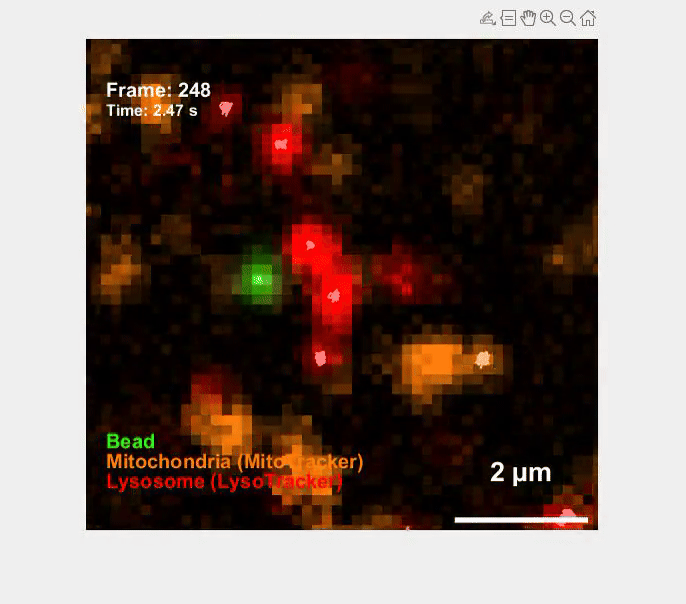
Mechanics
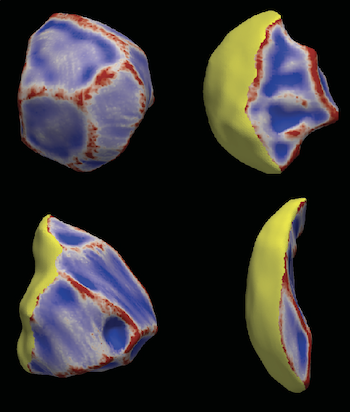
Geometry
3. Multiscale Perturbation & Predictive Modeling
Correlation alone cannot prove causation. We implement targeted perturbations across scales—CRISPR–Cas9 edits, optogenetic modulation, precise mechanical manipulations, and synthetic signaling circuits—coupled to live imaging. These interventions feed into machine-learning-driven models to chart feedback loops, test mechanistic hypotheses, and build predictive frameworks of how local interactions scale up to tissue-level form.
4. 3D In Vitro Model Systems for Developmental Reconstruction
To overcome in vivo constraints and explore variation, we apply our toolkit to stem-cell–derived systems—embryoids, gastruloids, organoids, and synthetic embryos. This ground-up approach defines minimal biochemical, mechanical, and geometric inputs required for reproducible self-organization, maps the morphospace of multicellular forms, and advances tissue engineering for regenerative medicine.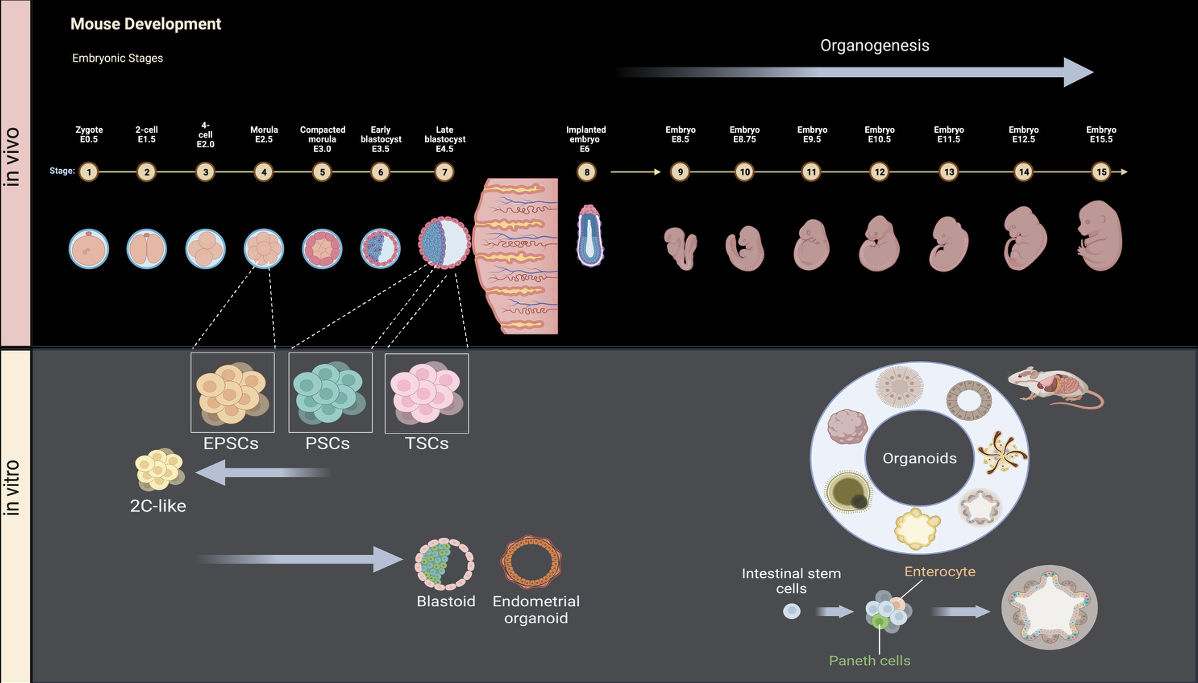
Selected Publications
Di Bella, D. J. *, Habibi, E. *, Stickels, R. R., Scalia, G., Brown, J., Yadollahpour, P., Yang, S. M., Abbate, C., Biancalani, T., Macosko, E. Z., Chen, F., Regev, A. & Arlotta, P. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature595, 554-559, doi:10.1038/s41586-021-03670-5 (2021).
Habibi, E. & Stunnenberg, H. G. Transcriptional and epigenetic control in mouse pluripotency: lessons from in vivo and in vitro studies. Curr Opin Genet Dev 46, 114-122, doi:10.1016/j.gde.2017.07.005 (2017).
von Meyenn, F. *, Iurlaro, M. *, Habibi, E. *, Liu, N. Q., Salehzadeh-Yazdi, A., Santos, F., Petrini, E., Milagre, I., Yu, M., Xie, Z., Kroeze, L. I., Nesterova, T. B., Jansen, J. H., Xie, H., He, C., Reik, W. & Stunnenberg, H. G. Impairment of DNA Methylation Maintenance Is the Main Cause of Global Demethylation in Naive Embryonic Stem Cells. Mol Cell 62, 848-861, doi:10.1016/j.molcel.2016.04.025 (2016).
Novakovic, B. *, Habibi, E. *, Wang, S. Y. *, Arts, R. J. W., Davar, R., Megchelenbrink, W., Kim, B., Kuznetsova, T., Kox, M., Zwaag, J., Matarese, F., van Heeringen, S. J., Janssen-Megens, E. M., Sharifi, N., Wang, C., Keramati, F., Schoonenberg, V., Flicek, P., Clarke, L., Pickkers, P., Heath, S., Gut, I., Netea, M. G., Martens, J. H. A., Logie, C. & Stunnenberg, H. G. beta-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell 167, 1354-1368 e1314, doi:10.1016/j.cell.2016.09.034 (2016).
Habibi, E., Brinkman, A. B., Arand, J., Kroeze, L. I., Kerstens, H. H., Matarese, F., Lepikhov, K., Gut, M., Brun-Heath, I., Hubner, N. C., Benedetti, R., Altucci, L., Jansen, J. H., Walter, J., Gut, I. G., Marks, H. & Stunnenberg, H. G. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13, 360-369, doi:10.1016/j.stem.2013.06.002 (2013).