Labs and Groups
Picketts Lab
Picketts Lab
Team Leader

David Picketts
Senior Scientist, Regenerative MedicineWhat We Do
Epigenetic Regulation in Brain Development and Disease Pathology
Disease gene identification for neurodevelopmental disorders has demonstrated that a large number encode epigenetic regulators. Amongst these are the ATP-dependent chromatin remodeling proteins that utilize the energy from ATP-hydrolysis to remove, remodel, or replace nucleosomes. Research in Dr. Picketts’ laboratory primarily focuses on understanding how aberrant chromatin remodeling affects brain development, cognition, and brain repair. Determining the genes and developmental pathways affected by these aberrant chromatin remodelers is critical to understanding human disease and for the generation of novel therapeutics.
Ongoing Projects:
1. Defining ATRX functions in neural development
We generated the first mouse models of the ATR-X syndrome and demonstrated a role for ATRX in mediating replication stress in the dividing progenitor cell population. We continue to investigate ATRX interacting proteins involved in mediating replication stress using both in vitro and in vivo model systems. A prominent unanswered question is what does ATRX do in a post-mitotic neuron? To address this question, we study the role of ATRX in the retina where it has importance cell non-autonomous roles and in a novel mouse model that presents with prominent features observed in human patients. Current projects with these animal models should highlight the molecular mechanisms underlying neuronal functions and learning deficits.
2. Mammalian ISWI proteins as mediators of brain size
The ISWI proteins, SNF2H and SNF2L co-regulate the expression of key neural developmental TFs (eg. Foxg1, En1) to regulate brain size. Snf2h ablation promotes progenitor differentiation while Snf2l loss enhances progenitor expansion to increase neuronal output. While this was thought to result from the actions of Snf2h- or Snf2l-specific complexes, recent studies have indicated that Snf2h and Snf2l are interchangeable. As such, differences in intrinsic remodeling activity of Snf2h versus Snf2l, and/or the temporal expression of different complexes may be key determinants. Current projects focus on characterizing the roles of individual complexes (eg. NuRF) through the analysis of partner proteins, and on genomic studies to define the co-regulated genes and remodeling alterations that drive the balance between proliferation and differentiation.
3. Mutations in PHF6 cause the Borjeson-Forssman-Lehmann (BFLS) syndrome
BFLS is caused by mutations in the PHF6 gene. PHF6 has two PHD domains and localizes to both the nucleus and the nucleolus. We have shown that it interacts with the NURD remodeling complex within the nucleus and regulates rDNA expression with the nucleolus. We have recently generated transgenic mice harboring the common human mutation (p.R342X) which will help us delineate its role in neocortical development and cognition.
4. VGF Neuropeptides as Mediators of Neuronal Damage
We identified VGF as an upregulated gene in exercising Snf2h conditional knockout mice. The upregulation of VGF promoted the survival of these animals, in part, by stimulating the clearance of cellular debris and the remyelination of neuronal axons. This work indicated that VGF should be examined as a potential therapeutic to promote brain recovery. Ongoing projects are exploring the role of VGF in promoting stroke recovery and for enhancing remyelination in models of Multiple Sclerosis.
Disease gene identification for neurodevelopmental disorders has demonstrated that a large number encode epigenetic regulators. Amongst these are the ATP-dependent chromatin remodeling proteins that utilize the energy from ATP-hydrolysis to remove, remodel, or replace nucleosomes. Research in Dr. Picketts’ laboratory primarily focuses on understanding how aberrant chromatin remodeling affects brain development, cognition, and brain repair. Determining the genes and developmental pathways affected by these aberrant chromatin remodelers is critical to understanding human disease and for the generation of novel therapeutics.
Ongoing Projects:
1. Defining ATRX functions in neural development
We generated the first mouse models of the ATR-X syndrome and demonstrated a role for ATRX in mediating replication stress in the dividing progenitor cell population. We continue to investigate ATRX interacting proteins involved in mediating replication stress using both in vitro and in vivo model systems. A prominent unanswered question is what does ATRX do in a post-mitotic neuron? To address this question, we study the role of ATRX in the retina where it has importance cell non-autonomous roles and in a novel mouse model that presents with prominent features observed in human patients. Current projects with these animal models should highlight the molecular mechanisms underlying neuronal functions and learning deficits.
2. Mammalian ISWI proteins as mediators of brain size
The ISWI proteins, SNF2H and SNF2L co-regulate the expression of key neural developmental TFs (eg. Foxg1, En1) to regulate brain size. Snf2h ablation promotes progenitor differentiation while Snf2l loss enhances progenitor expansion to increase neuronal output. While this was thought to result from the actions of Snf2h- or Snf2l-specific complexes, recent studies have indicated that Snf2h and Snf2l are interchangeable. As such, differences in intrinsic remodeling activity of Snf2h versus Snf2l, and/or the temporal expression of different complexes may be key determinants. Current projects focus on characterizing the roles of individual complexes (eg. NuRF) through the analysis of partner proteins, and on genomic studies to define the co-regulated genes and remodeling alterations that drive the balance between proliferation and differentiation.
3. Mutations in PHF6 cause the Borjeson-Forssman-Lehmann (BFLS) syndrome
BFLS is caused by mutations in the PHF6 gene. PHF6 has two PHD domains and localizes to both the nucleus and the nucleolus. We have shown that it interacts with the NURD remodeling complex within the nucleus and regulates rDNA expression with the nucleolus. We have recently generated transgenic mice harboring the common human mutation (p.R342X) which will help us delineate its role in neocortical development and cognition.
4. VGF Neuropeptides as Mediators of Neuronal Damage
We identified VGF as an upregulated gene in exercising Snf2h conditional knockout mice. The upregulation of VGF promoted the survival of these animals, in part, by stimulating the clearance of cellular debris and the remyelination of neuronal axons. This work indicated that VGF should be examined as a potential therapeutic to promote brain recovery. Ongoing projects are exploring the role of VGF in promoting stroke recovery and for enhancing remyelination in models of Multiple Sclerosis.
Research Activities
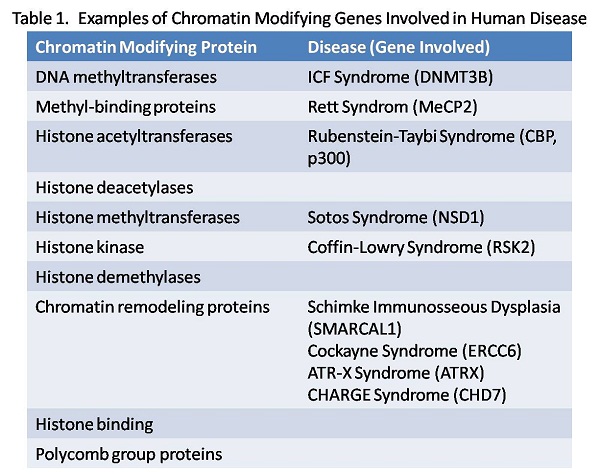
Chromatin remodeling proteins and XLID: Chromatin remodeling proteins play a dynamic role in the regulation of gene expression through the alteration of nucleosome structure (histone acetylation, phosphorylation and methylation) or the ATP dependent repositioning of nucleosomes (SWI/SNF complex, ISWI). Their involvement in genetic disease was established by our cloning of the ATRX gene, a novel SWI/SNF family member that is mutated in a severe X-linked intellectual disability syndrome usually associated with alpha thalassemia. This paradigm has since been extended to include genes encoding almost every type of chromatin modifying protein (see Table 1). We continue to study the role of these proteins in neural development to understand how they contribute to disease pathogenesis. Several current projects are described below.
1. ATRX function and the ATR-X syndrome: We make use of multiple Cre drivers to conditionally inactivate Atrx (cKO) in mouse tissues involved in the human syndrome. We have made use of a muscle cKO to define a role for Atrx in replication and/or maintenance of heterochromatin. We identified a cell non-autonomous role for Atrx in the developing retina that affects the survival of inter-neurons. Finally, we are defining the role of Atrx in the developing forebrain. Each of these projects is ongoing and continues to provide fundamental knowledge on the global and cell type specific roles of Atrx.

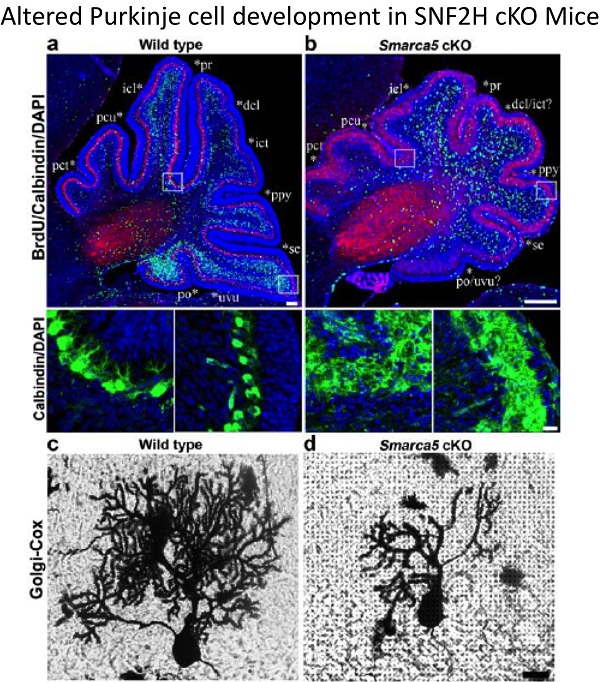
2. Dual functions of the ISWI proteins SNF2H and SNF2L: To extend our analysis of the role of chromatin remodeling proteins in neural development we have characterized the human and murine SNF2H and SNF2L genes. SNF2H is expressed in proliferating neuronal cell populations whereas SNF2L is expressed predominantly in differentiating and/or maturing neurons. Mice inactivated for SNF2L have enlarged brains due to enhanced proliferation whereas mice deficient for SNF2H have proliferative defects and underdeveloped brains. Current lab work aims to understand how these genes regulate common master neuronal developmental homeotic genes to control the size and patterning of specific brain structures.
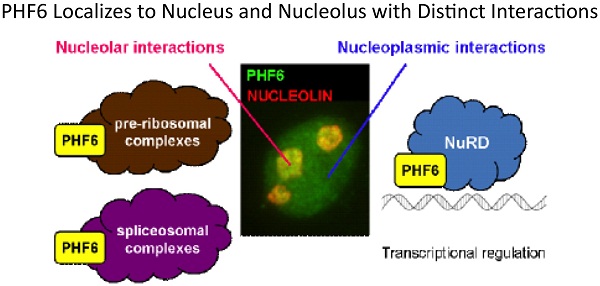
3. PHF6, a causative gene for two distinct diseases: PHF6 contains two PHD-like chromatin domains and is the cause of Borjeson-Forssman-Lehmann syndrome, an XLID, and is also mutated in a significant proportion of T-cell acute lymphocytic leukemia. We undertook a biochemical approach to identify its interacting partners to help elucidate its role in such disparate diseases. We determined that PHF6 interacts with the NuRD complex and future work is aimed at identifying specific targets genes regulated by the PHF6-NuRD complex.

1. ATRX function and the ATR-X syndrome: We make use of multiple Cre drivers to conditionally inactivate Atrx (cKO) in mouse tissues involved in the human syndrome. We have made use of a muscle cKO to define a role for Atrx in replication and/or maintenance of heterochromatin. We identified a cell non-autonomous role for Atrx in the developing retina that affects the survival of inter-neurons. Finally, we are defining the role of Atrx in the developing forebrain. Each of these projects is ongoing and continues to provide fundamental knowledge on the global and cell type specific roles of Atrx.

2. Dual functions of the ISWI proteins SNF2H and SNF2L: To extend our analysis of the role of chromatin remodeling proteins in neural development we have characterized the human and murine SNF2H and SNF2L genes. SNF2H is expressed in proliferating neuronal cell populations whereas SNF2L is expressed predominantly in differentiating and/or maturing neurons. Mice inactivated for SNF2L have enlarged brains due to enhanced proliferation whereas mice deficient for SNF2H have proliferative defects and underdeveloped brains. Current lab work aims to understand how these genes regulate common master neuronal developmental homeotic genes to control the size and patterning of specific brain structures.

3. PHF6, a causative gene for two distinct diseases: PHF6 contains two PHD-like chromatin domains and is the cause of Borjeson-Forssman-Lehmann syndrome, an XLID, and is also mutated in a significant proportion of T-cell acute lymphocytic leukemia. We undertook a biochemical approach to identify its interacting partners to help elucidate its role in such disparate diseases. We determined that PHF6 interacts with the NuRD complex and future work is aimed at identifying specific targets genes regulated by the PHF6-NuRD complex.

Selected Publications
Gibbons, RJ, Picketts, DJ, and Higgs, DR. (1995) X-linked mental retardation associated with a thalassaemia (ATR-X syndrome) results from mutations in a putative global transcriptional regulator. Cell 80, 837-845.
Barak, O., Lazzaro, MA., Lane, WS., Speicher DW., Picketts, DJ., Shiekhattar R. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. (2003) 22:6089-6100.
Bérubé, NG, Jagla M, Vanderluit, JL, Garrick, D, Gibbons, RJ, Higgs, DR, Slack, RS, and Picketts, DJ. The chromatin remodeling protein ATRX is critical for neuronal survival during corticogenesis. (2005) J. Clin. Invest. 115: 258-267.
Yip, DJ, Corcoran, CP, DeMaria, A, Rennick, S, Rudnicki, MA, Messier, C, and Picketts DJ. Snf2l regulates Foxg1-dependent progenitor cell expansion in the developing brain. (2012) Dev Cell. 2012 Apr 17;22(4):871-8.
Alvarez-Saavedra, M., Lagali, P., Yan, K., Mears, A., De Repentigny, Y., Hashem, E., Wallace, V.A., Kothary, R., Stopka, T., Skoultchi, A.I., and Picketts, D.J. Snf2h mediates histone dynamics to control cerebellar development and function. Nature Comm (2014) 5:4181
Alvarez-Saavedra M., De Repentigny Y., Yang D., O'Meara R., Yan K., Racacho L., Ioshikhes I., Parks RJ., Kothary R., Picketts, D.J. (2016) Voluntary Running Triggers VGF-Mediated Oligodendrogenesis to Prolong the Lifespan of Snf2h-Null Ataxic Mice. Cell Rep. 17: 862-875
Barak, O., Lazzaro, MA., Lane, WS., Speicher DW., Picketts, DJ., Shiekhattar R. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. (2003) 22:6089-6100.
Bérubé, NG, Jagla M, Vanderluit, JL, Garrick, D, Gibbons, RJ, Higgs, DR, Slack, RS, and Picketts, DJ. The chromatin remodeling protein ATRX is critical for neuronal survival during corticogenesis. (2005) J. Clin. Invest. 115: 258-267.
Yip, DJ, Corcoran, CP, DeMaria, A, Rennick, S, Rudnicki, MA, Messier, C, and Picketts DJ. Snf2l regulates Foxg1-dependent progenitor cell expansion in the developing brain. (2012) Dev Cell. 2012 Apr 17;22(4):871-8.
Alvarez-Saavedra, M., Lagali, P., Yan, K., Mears, A., De Repentigny, Y., Hashem, E., Wallace, V.A., Kothary, R., Stopka, T., Skoultchi, A.I., and Picketts, D.J. Snf2h mediates histone dynamics to control cerebellar development and function. Nature Comm (2014) 5:4181
Alvarez-Saavedra M., De Repentigny Y., Yang D., O'Meara R., Yan K., Racacho L., Ioshikhes I., Parks RJ., Kothary R., Picketts, D.J. (2016) Voluntary Running Triggers VGF-Mediated Oligodendrogenesis to Prolong the Lifespan of Snf2h-Null Ataxic Mice. Cell Rep. 17: 862-875